This guide explains how to stream EEG data from eego™ devices into BrainFlow using Python. It includes setup steps and how to adapt sample scripts for ANT Neuro amplifiers. BrainFlow offers real-time data visualization and analysis but is supported by its own community.
AIM
This document illustrates how data can be streamed into the BrainFlow platform using the eego™ SDK. BrainFlow is a library that enables acquisition, plotting and analysis of EEG data from a variety of systems. The next sections will walk the reader through the BrainFlow platform and its intended use, provide step-by-step installation instructions for Python version 3.1 and above. Finally, we include some plots of the data that is readily available with an exemplar script in the BrainFlow package.
Waiver:
The instructions for installing and adapting of the BrainFlow toolbox are examples only and may be applicable only for the specific version tested. ANT Neuro cannot provide support for the BrainFlow toolbox itself. For support, please address the BrainFlow community.
BACKGROUND
BrainFlow is a library intended to obtain, parse and analyze EEG, EMG, ECG and other kinds of data from biosensors⁴. It is a useful plotting and analysis tool to employ when working with just the eego™ SDK. The BrainFlow platform is compatible with our EE-4XX and EE-2XX eego™ amplifiers⁵ and can makes real-time signal visualization and filtering easier for SDK users⁶.
The primary focus of the developers of BrainFlow is to deliver an easy to use, powerful Application Programming Interface (API) for data acquisition and signal processing⁴. All features on the platform are available for free and distributed under the MIT license⁷. Synthetic boards for eego™ amplifiers are useful development tools that can also be employed by users⁵.
INSTALLATION INSTRUCTIONS TO SETUP BRAINFLOW
Step Number | Description | Command Line(s) |
1. Download BrainFlow | Download BrainFlow from BrainFlow Github⁸ and store the folder under the filepath ‘C:\Users\Username\’. | |
2. Download and Install an open-source Python distribution platform such as Anaconda to run Python versions 3.1 and above | Python version 3.8.13 was used for this testing, Python version 3.1 or higher needs to be used for the Brainflow platform.⁹ | python –version Typing out this command on the Anaconda Prompt window will give you the installed Python version. |
3. Create a new Python environment | Open the Anaconda Prompt application to create a new Python BrainFlow environment
| conda create --name env name python=3.8 (e. g. conda create --name brainflowpython=3.8) |
4. Activate the Brainflow environment | Activate this newly created BrainFlow environment via Anaconda Prompt | conda activate env name |
5. Install BrainFlow | Install the latest BrainFlow release from PyPI packages with the following command | python -m pip install brainflow |
6. Build the CMake project | Change the location to the directory where BrainFlow folder is stored and run the following command to build the CMake project (no additional options need to be set by users) | python -m pip install cmake
cd tools
python build.py |
SAMPLE SCRIPT TO PLOT EEG DATA IN REAL-TIME
In this section, we focus on one example Python script available to all users after completing the installation steps above. Quite a number of sample scripts for the BrainFlow application are available via the BrainFlow website⁶, for different packages such as Matlab, Python, Java, R etc. Our focus here was the sample script found under the Python Packages subfolder, titled ‘plot_real_time.py’. The file path on your local PC where this can be found after installation is successfully completed is: “…\brainflow-master\python-package\examples\plot_real_time”.
All scripts can be run via Python IDEs such as Spyder or PyCharm. The Brainflow platform supports hardware from multiple manufacturers and assigns a Board ID to each device type. All such supported boards can be found under: https://brainflow.readthedocs.io/en/stable/SupportedBoards.html⁵.
To choose the ANT Neuro boards or amplifier type in the BoardShim constructor, please specify according to the amplifier type or the product code seen on the back label of the amplifier. For example, when using the eego™ mini amplifier, with the product code EE-411, choose, ANT_NEURO_EE_411_BOARD (board id 25).
Within the ‘plot_real_time.py’ script, the section below would have to be adapted when using a specific ANT Neuro board ID or amplifier type:
In line 124, under section ‘def main()’, the default is:
parser.add_argument('--board-id', type=int, help='board id, check docs to get a list of supported boards', required=False, default=BoardIds.SYNTHETIC_BOARD)
Update this line to include the amplifier type in use, for example:
parser.add_argument('--board-id', type=int, help='board id, check docs to get a list of supported boards', required=False, default=BoardIds.ANT_NEURO_EE_411_BOARD)
Figure 1 is a direct output of this script. It was recorded with the eego™ mini amplifier and an 8-channel waveguard™touch cap:
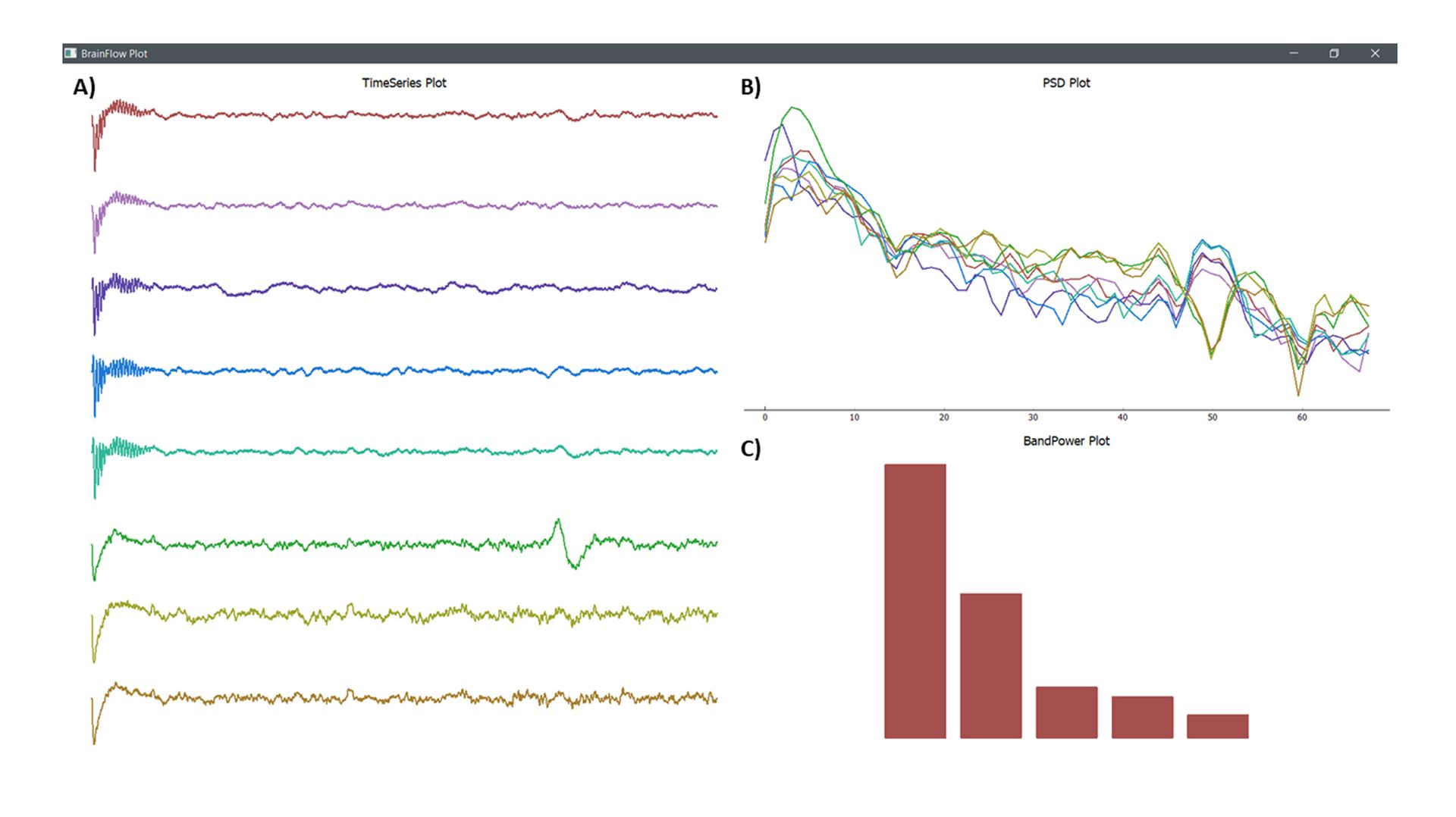
Figure 1: Plots generated by the sample script.
Figure 1.A: Time series EEG data (in µV) retrieved from an 8 channel eego™ mini amplifier using an 8-channel waveguard™ touch cap. Figure 1.B: Power spectral density (PSD) plot depicts the frequency content of the signal or the power distribution of the EEG in the frequency domain. Figure 1.C: The Band power barplot displays the contribution of a given frequency band to overall power of the signal in percentages (%).
Data Format Description – Unit of Measures
(Please also see source under Brainflow website)¹⁰
- For EEG channels BrainFlow returns µV wherever possible.
- BrainFlow uses UNIX timestamps, this counter starts at the Unix Epoch on January 1st, 1970 at UTC. Precision is in the microsecond range, but for some boards timestamps are generated on PC side as soon as packages have been received.
LIMITATIONS
- Online processing and filtering times might be longer when the channel count increases.
- The script variables can be changed to find the right filter values for your setup.
- Using the sample scripts require careful consideration as most functions are not editable to the user. Users can build on the sample scripts material to have the final plots as desired.
CONCLUSION
The BrainFlow open-source software solution used in combination with our eego™ SDK is an excellent tool for developing pipelines for real-time EEG data acquisition and preprocessing. Starting from BrainFlow release 5.0.0onwards, there is an option to load some Open Neural Network Exchange (ONNX) runtime models into BrainFlow11. ONNX is an open format for machine learning (ML) models which enables interchange of models between various ML frameworks and tools. This update would be especially helpful for the Machine Leaning community. We advise all users to continue exploring example scripts that are part of the Brainflow package. Adaptation of variables such as the default filtering values, adding axes and legends to the plots to further improve real-time plotting and visualization could be useful additions to the sample scripts. Finally, we would like to mention that the Brainflow slack channel continues to actively support queries related to their platform and the main developers moderate this slack channel routinely.
REFERENCES
- Anaconda | The World’s Most Popular Data Science Platform.https://www.anaconda.com/
- CMake. Accessed August 29, 2022. https://cmake.org/
- Visual Studio: IDE and Code Editor for Software Developers and Teams. https://visualstudio.microsoft.com/
- BrainFlow. https://brainflow.org/
- User API — BrainFlow documentation. https://brainflow.readthedocs.io/en/stable/UserAPI.html#python-api-reference
- Code Samples — BrainFlow documentation. https://brainflow.readthedocs.io/en/stable/Examples.html#python
- MIT License — BrainFlow documentation. https://brainflow.readthedocs.io/en/stable/License.html
- brainflow-dev/brainflow. https://github.com/brainflow-dev/brainflow
- Download Python. Python.org. https://www.python.org/downloads/
- Data Format Description — BrainFlow documentation. https://brainflow.readthedocs.io/en/stable/DataFormatDesc.html
- Parfenov A. BrainFlow with onnxruntime. https://brainflow.org/2022-06-09-onnx/
DISCLAIMER
All information provided in this document is intended as a summary only.
For detailed product related information please always consult the latest version of the respective product’s user manual. This document is not intended to replace the user documentation. For indications on individual feature certification status (clinical vs. research only) please refer to the aforementioned user documentation.
We have attempted to write this document as accurately as possible. However, mistakes are bound to occur, and we reserve the right to make changes whenever needed or whenever new information becomes available.
All product names and brand names in this document are trademarks or registered trademarks of their respective holders.
© Copyright 2022 eemagine Medical Imaging Solutions GmbH.
No part of this document may be photocopied or reproduced or transmitted in any way without prior written consent from eemagine Medical Imaging Solutions GmbH.